PKbioanalysis supports interactive dilution scheme generation.
library(PKbioanalysis)
#>
#> Attaching package: 'PKbioanalysis'
#> The following object is masked from 'package:stats':
#>
#> integrateLet’s generate fairly complex plate with multiple different standards, QC and DQCs.
x <- generate_96("Dilution integerity") |>
add_DB() |>
add_blank() |>
add_blank() |>
fill_scheme("h", lbound = 4, rbound = 11) |>
add_cs_curve(c(1, 3, 5, 10, 20, 50, 100, 200), rep =3) |>
fill_scheme("h", tbound = "D", bbound = "E", lbound = 1, rbound = 12) |>
add_QC(3, 90, 180, reg = T, n_qc = 6, qc_serial = F, group = "DrugA") |>
fill_scheme("h", lbound = 1, rbound = 4) |>
add_DQC(conc = 1000, 100, rep = 6) |>
fill_scheme("h", lbound = 1, rbound = 8) |>
add_cs_curve(c(1, 4, 6, 8, 15, 75, 150, 200)) |>
fill_scheme("h", tbound = "G", bbound = "H", lbound = 1, rbound = 8) |>
add_QC(3, 95, 190, reg = T, n_qc = 4, qc_serial = F, group = "DrugB") |>
fill_scheme("v", lbound = 9, rbound = 12, tbound = "F", bbound = "H") |>
add_DQC(conc = 1000, 10, rep = 6) |>
add_DQC(conc = 1000, 100, rep = 6)
plot(x)
#> Plate not registered. To register, use register_plate()
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_text()`).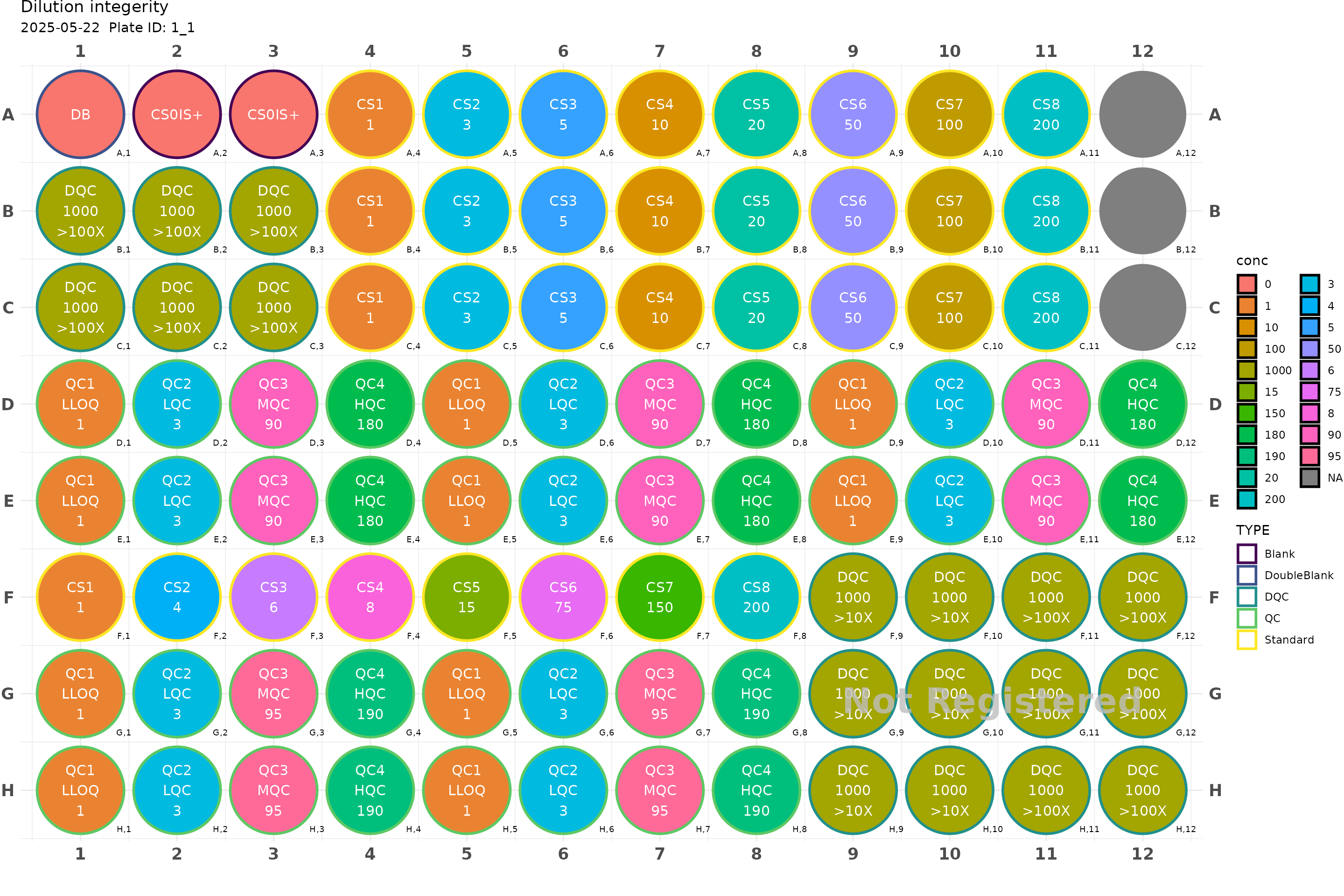
Accessing dilution scheme generation is done through the
study_app() function, which is same for historical plate
navigation and generation of injection sequence.
Make sure to select the plate you want to generate dilution scheme for. The last registered plate will be selected by default.
- From Generators, select
Dilution - Select the last dilution factor you want to use. The default is 10.
- Select the unit for the last 2 dilution steps. The default is
ng, but could use concentration units likeng/mLorng/uL. Also, you could delete the unit entirely. Just make sure the units can logically convert to each other. In other words, either all masses, all concentrations, or no units at all. - Select vial type. Either “Standard”, “QC”, or “DQC”.
- Finally, select which replicate you need to generat the dilution
scheme for and hit
Dilute
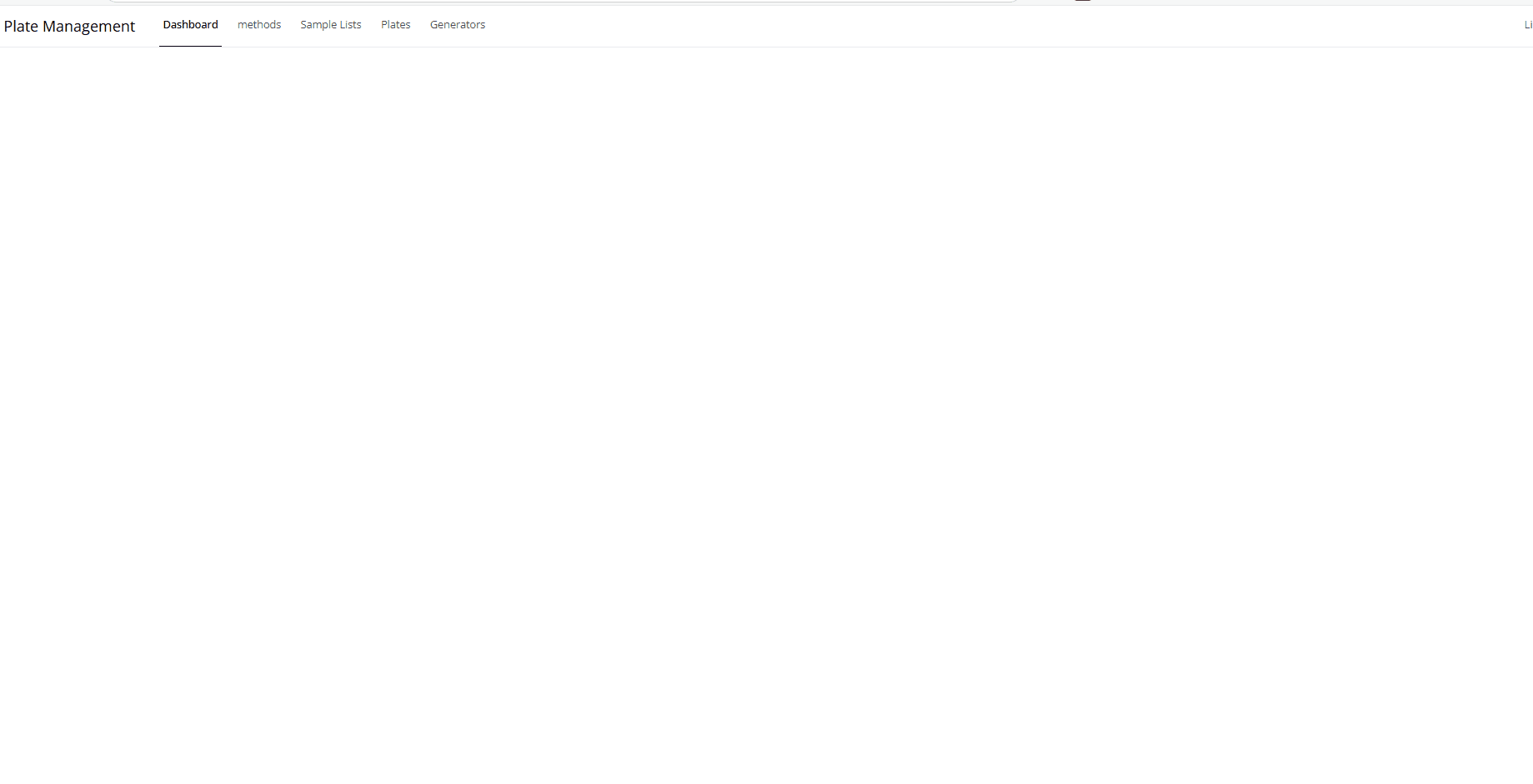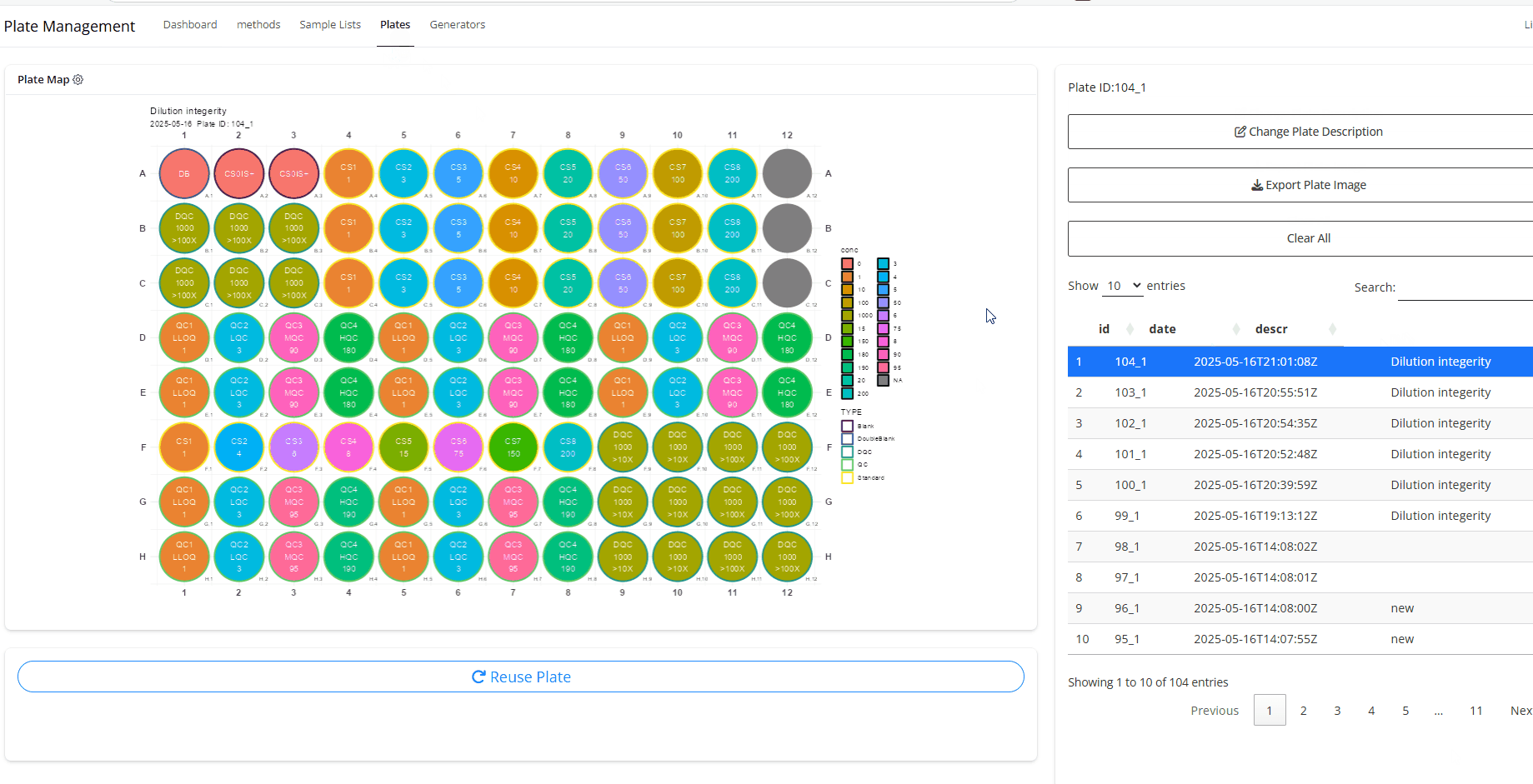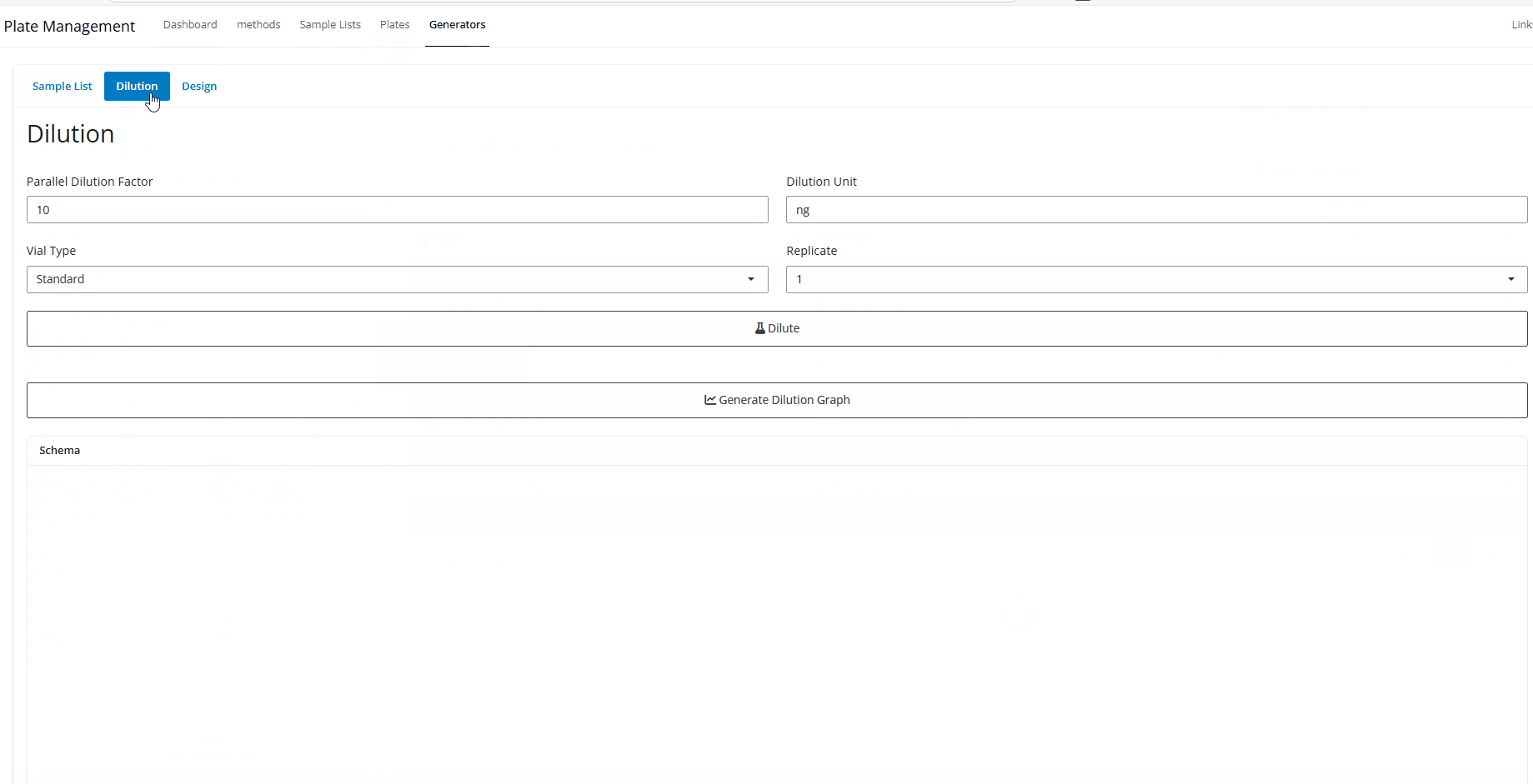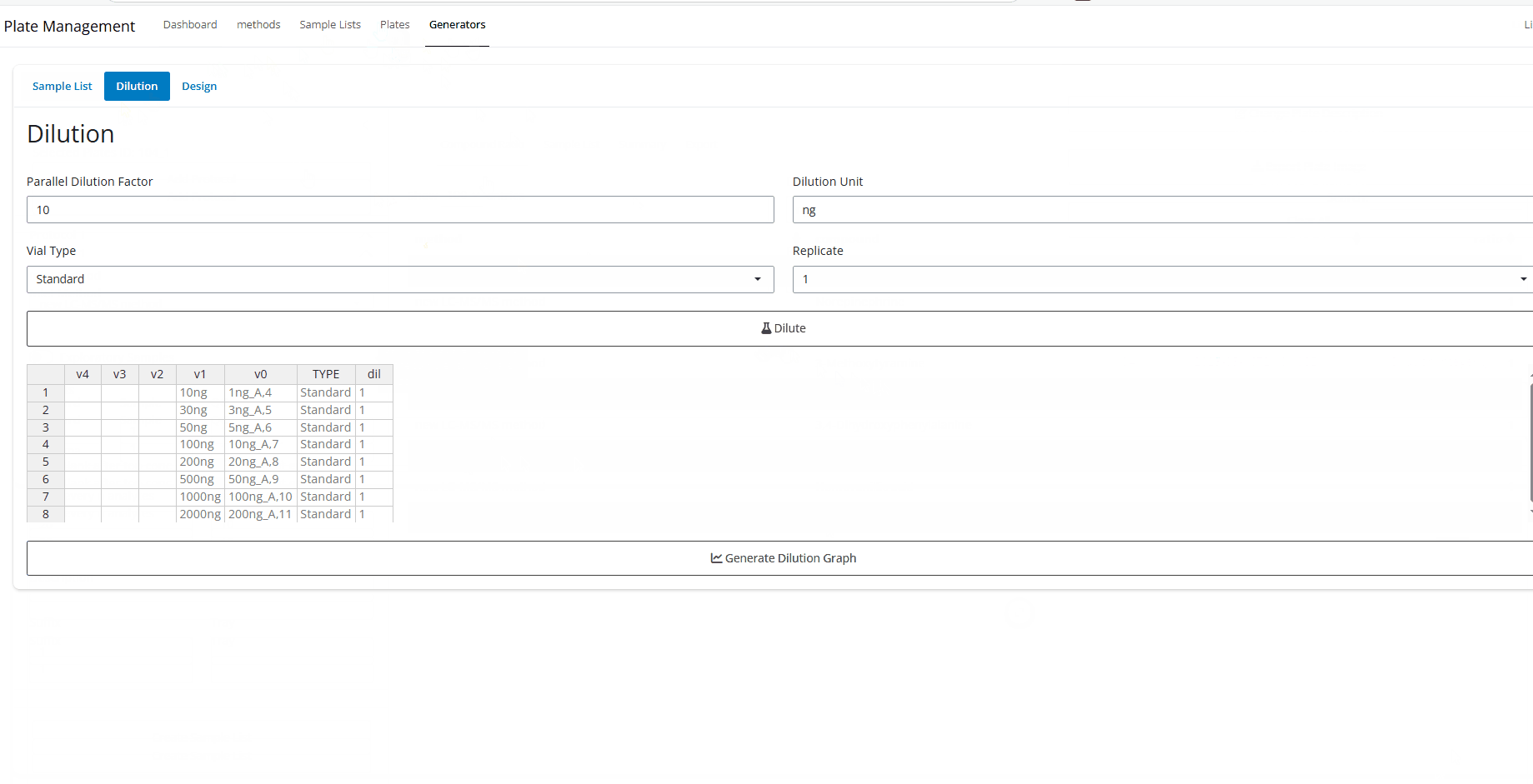
The table generated is interactive. It will also fill down if no concentration is specified. Again, must just units will be consistent.
It is highly recommend to keep dilution steps same and parallel as much as possible to mimize different error between aliquots.
Hit Generate Dilution Graph to get an interactive
dilution scheme.
Click on any non-terminal node, and specify the final volume needed. The calculation will be done automatically.
Warning: Currently, PKbioanalysis does not have database for dilution scheme. You must export the dilution scheme images by using
Exportbutton.
Currently, checking if DQC having starting concentration higher than ULOQ or if the dilution scheme include the specified dilution factor is no supported.
