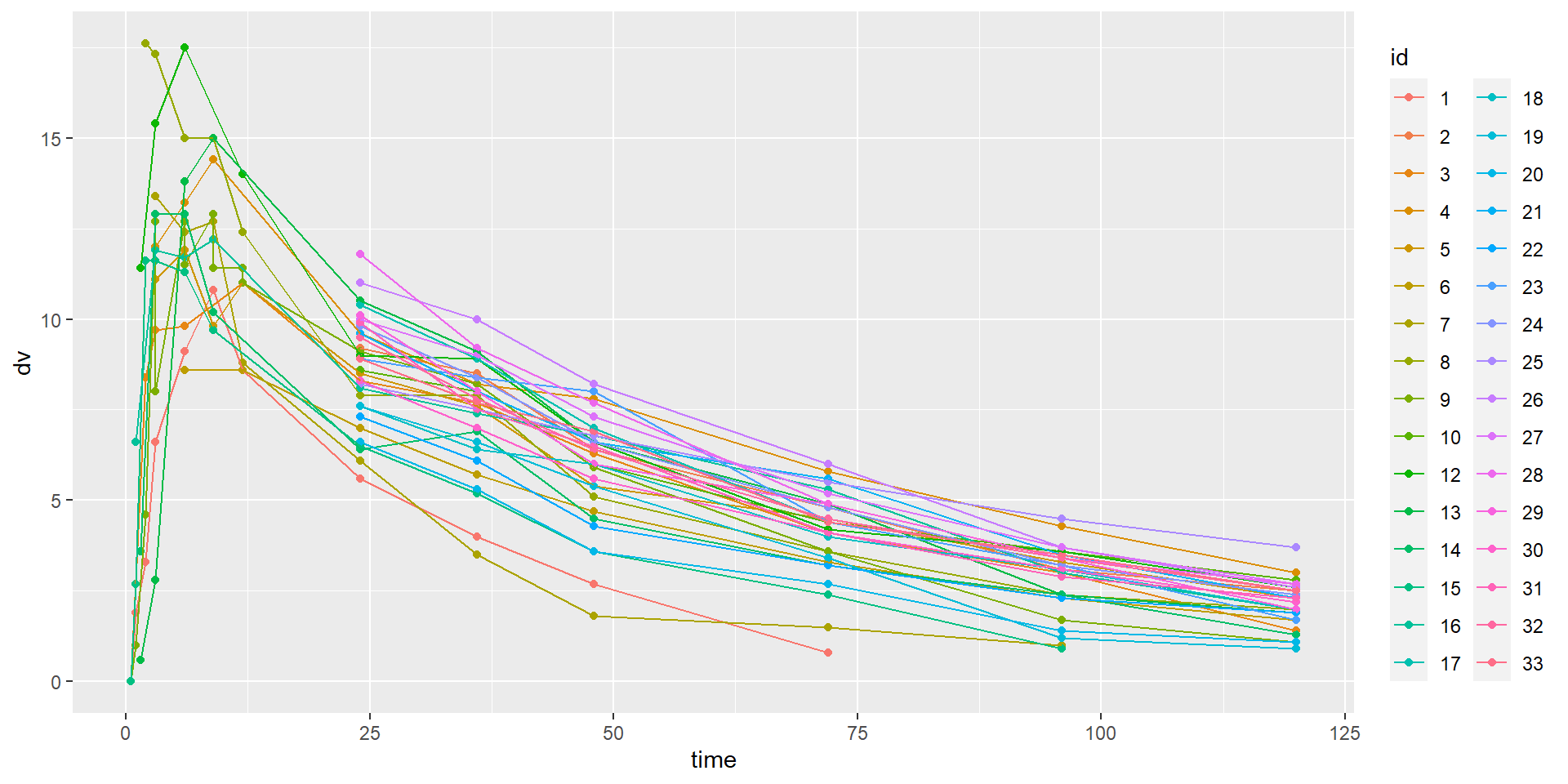
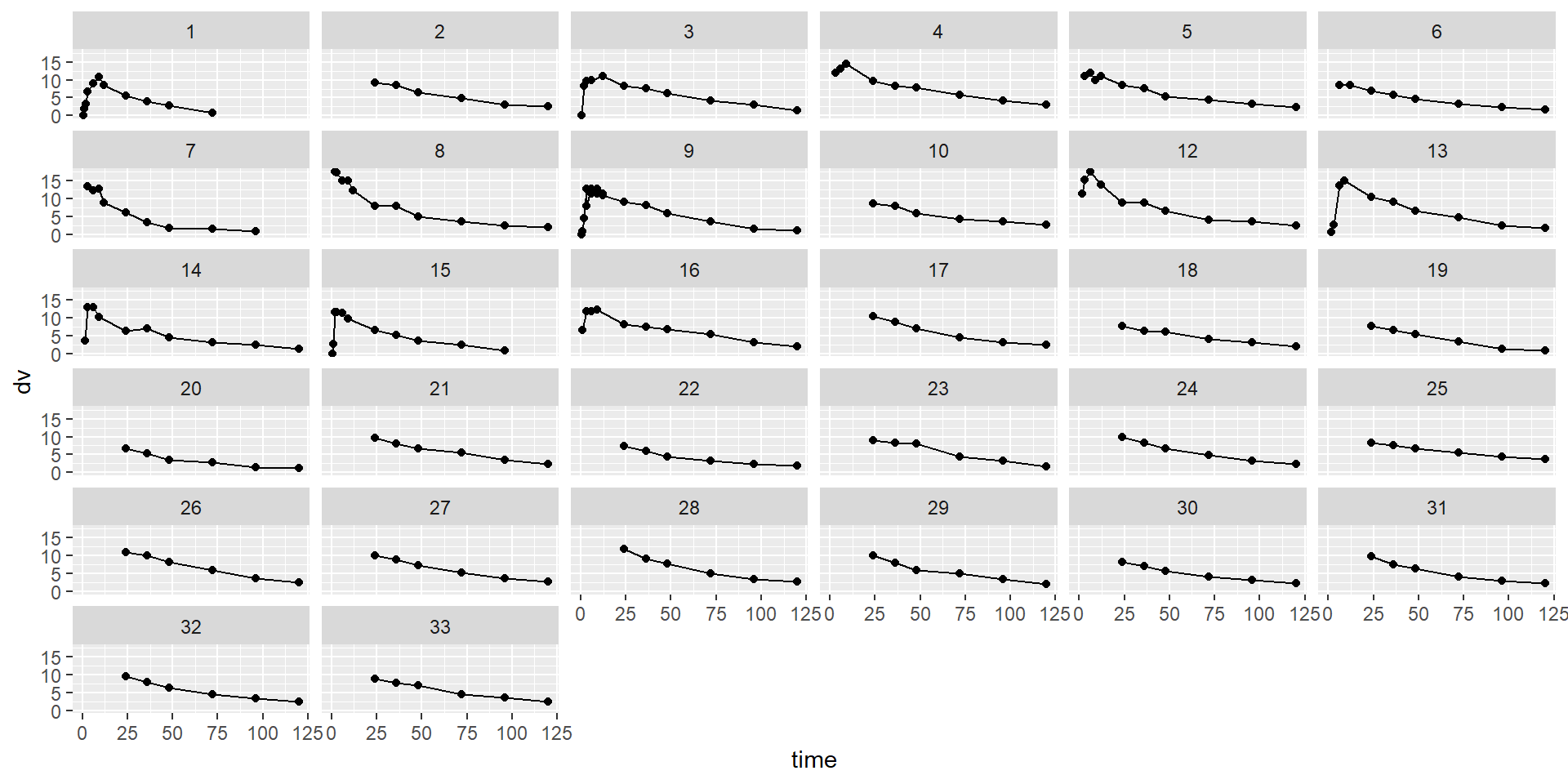
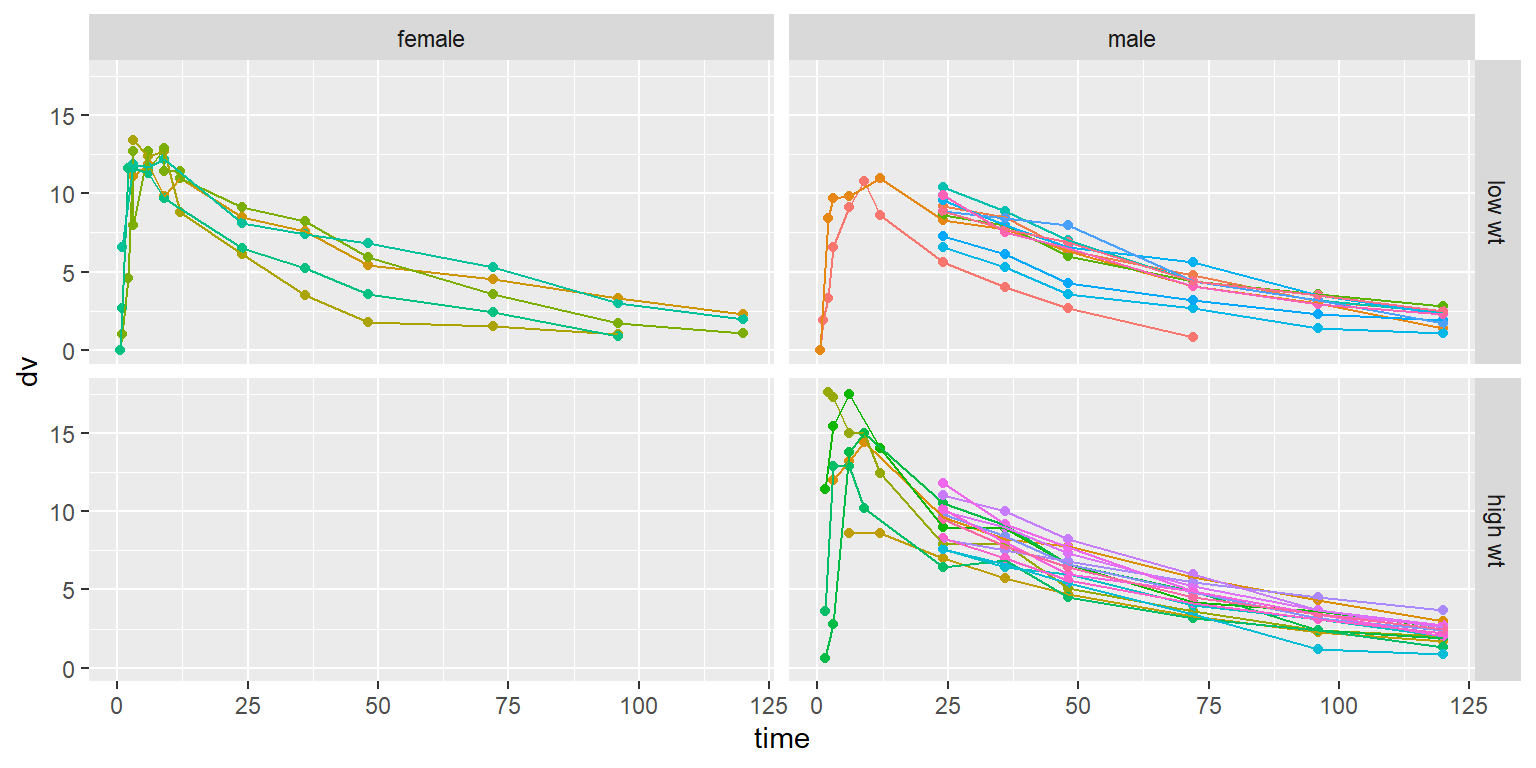
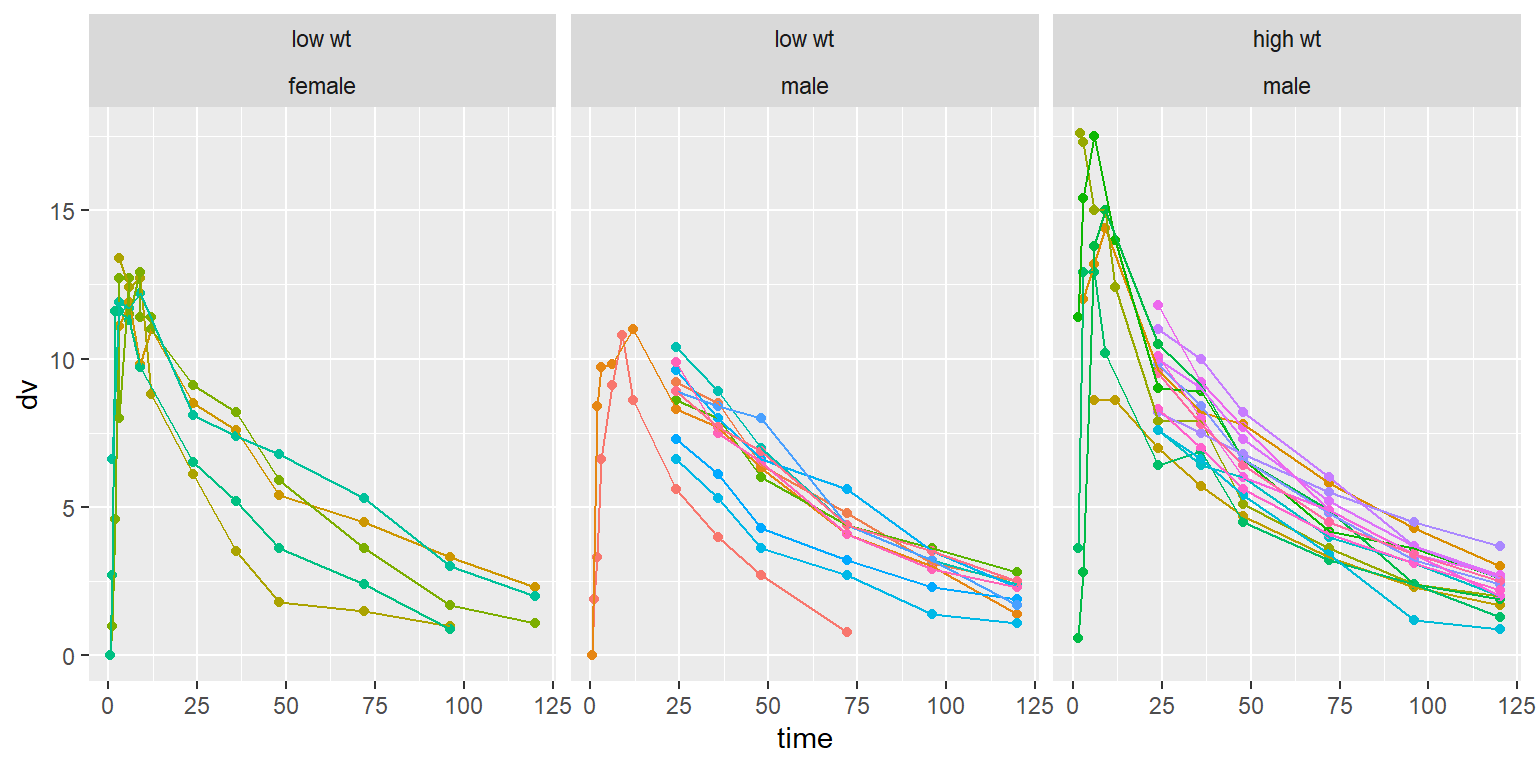
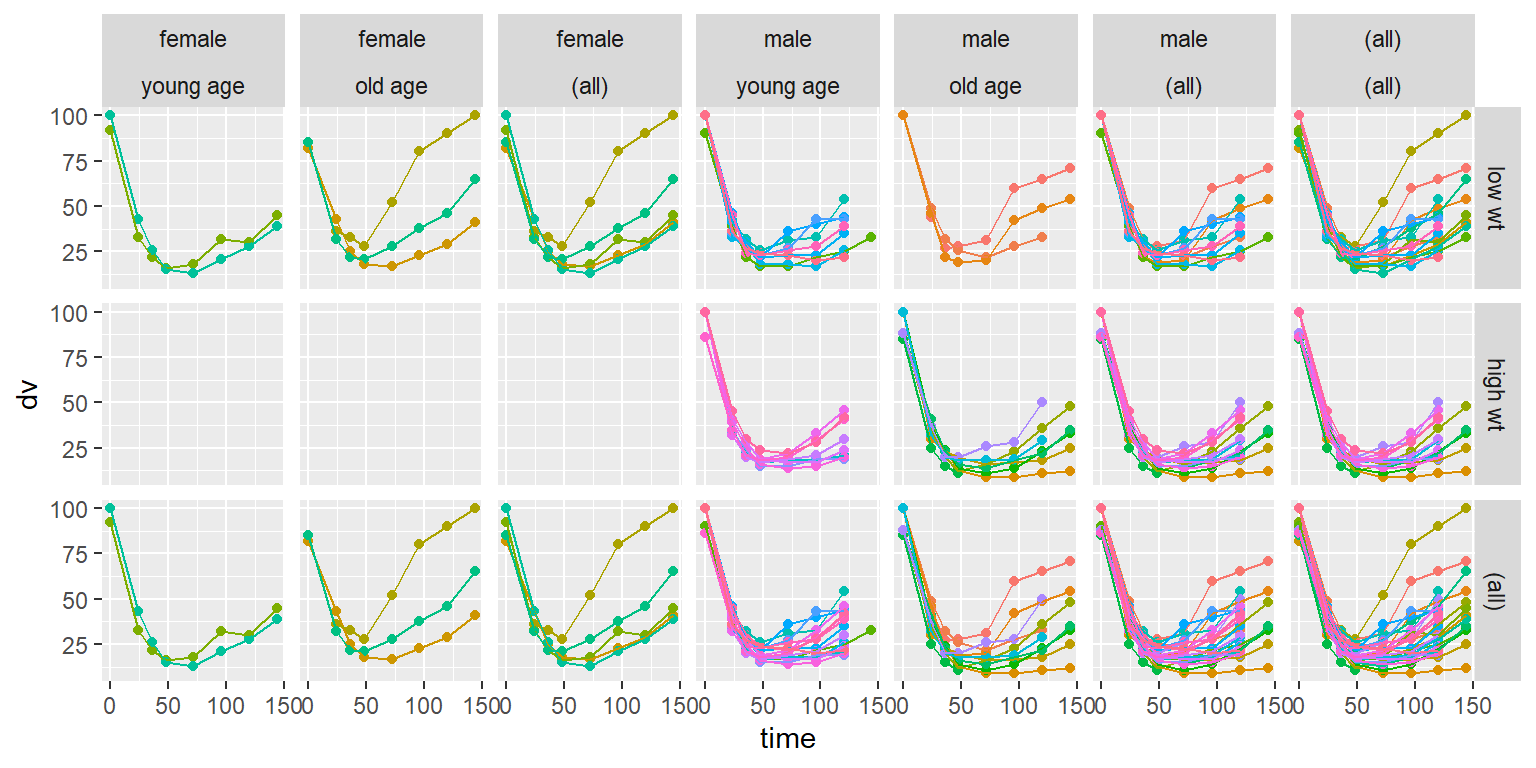
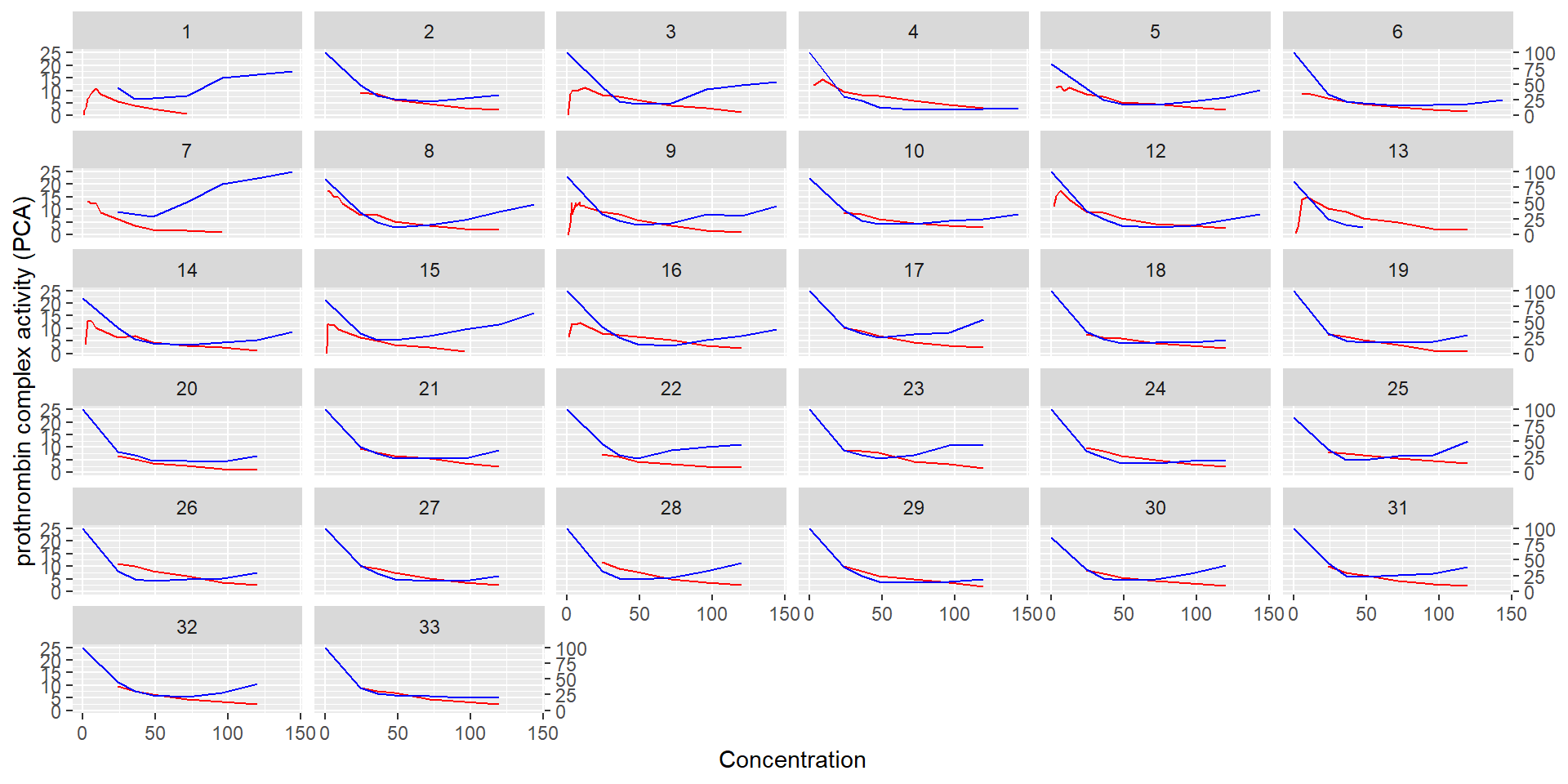
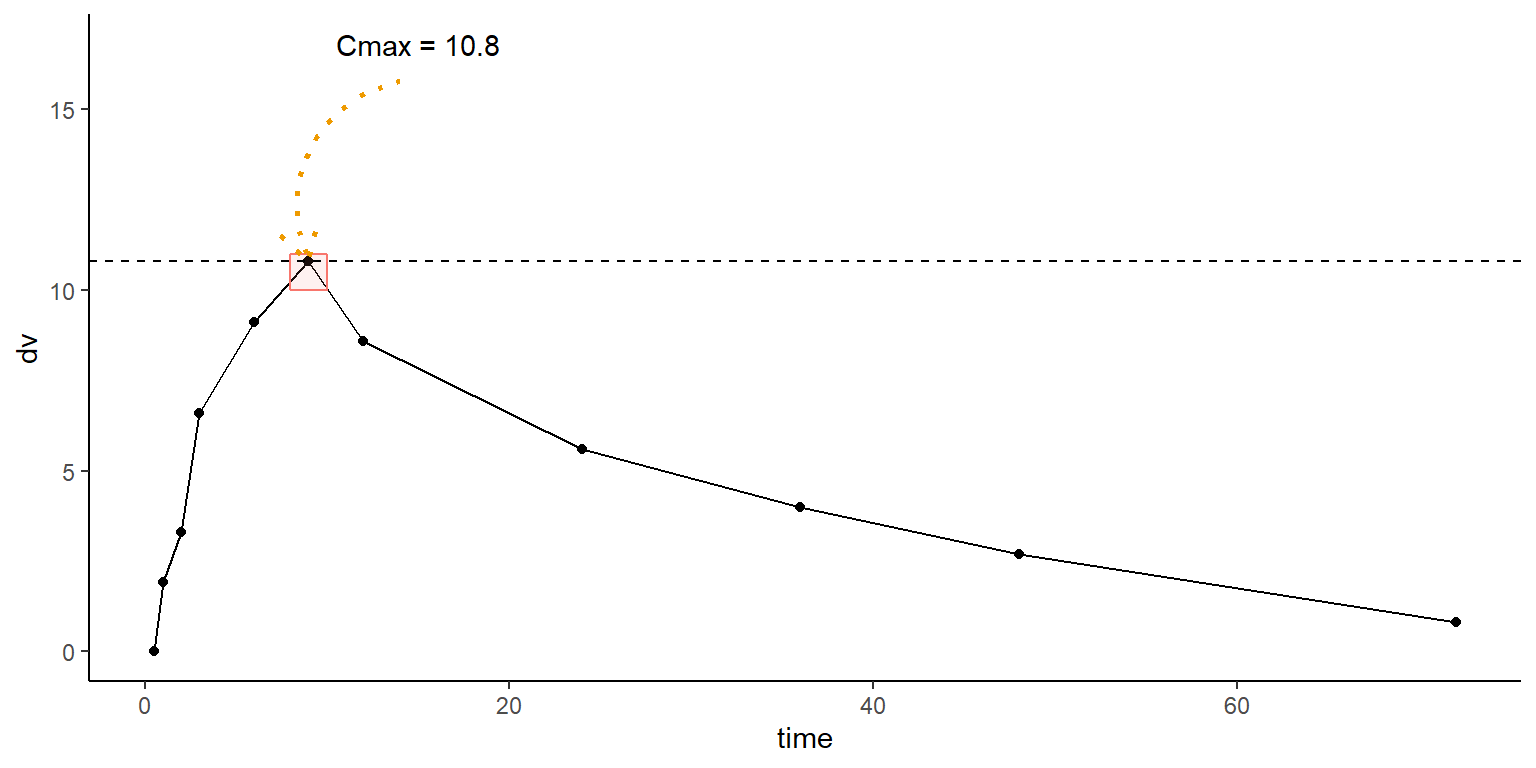
id time amt dv dvid evid wt age sex wt_cat age_cat
1 1 0.5 100 0.0 cp 0 66.7 50 male low wt old age
2 1 1.0 100 1.9 cp 0 66.7 50 male low wt old age
3 1 2.0 100 3.3 cp 0 66.7 50 male low wt old age
4 1 3.0 100 6.6 cp 0 66.7 50 male low wt old age
5 1 6.0 100 9.1 cp 0 66.7 50 male low wt old age
6 1 9.0 100 10.8 cp 0 66.7 50 male low wt old age id time amt dv dvid
9 : 17 Min. : 0.50 Min. : 60.0 Min. : 0.000 cp :251
1 : 11 1st Qu.: 18.00 1st Qu.: 88.5 1st Qu.: 3.200 pca: 0
3 : 11 Median : 36.00 Median :105.0 Median : 6.100
8 : 11 Mean : 50.07 Mean :103.1 Mean : 6.412
15 : 11 3rd Qu.: 72.00 3rd Qu.:117.0 3rd Qu.: 8.900
5 : 10 Max. :120.00 Max. :153.0 Max. :17.600
(Other):180
evid wt age sex wt_cat
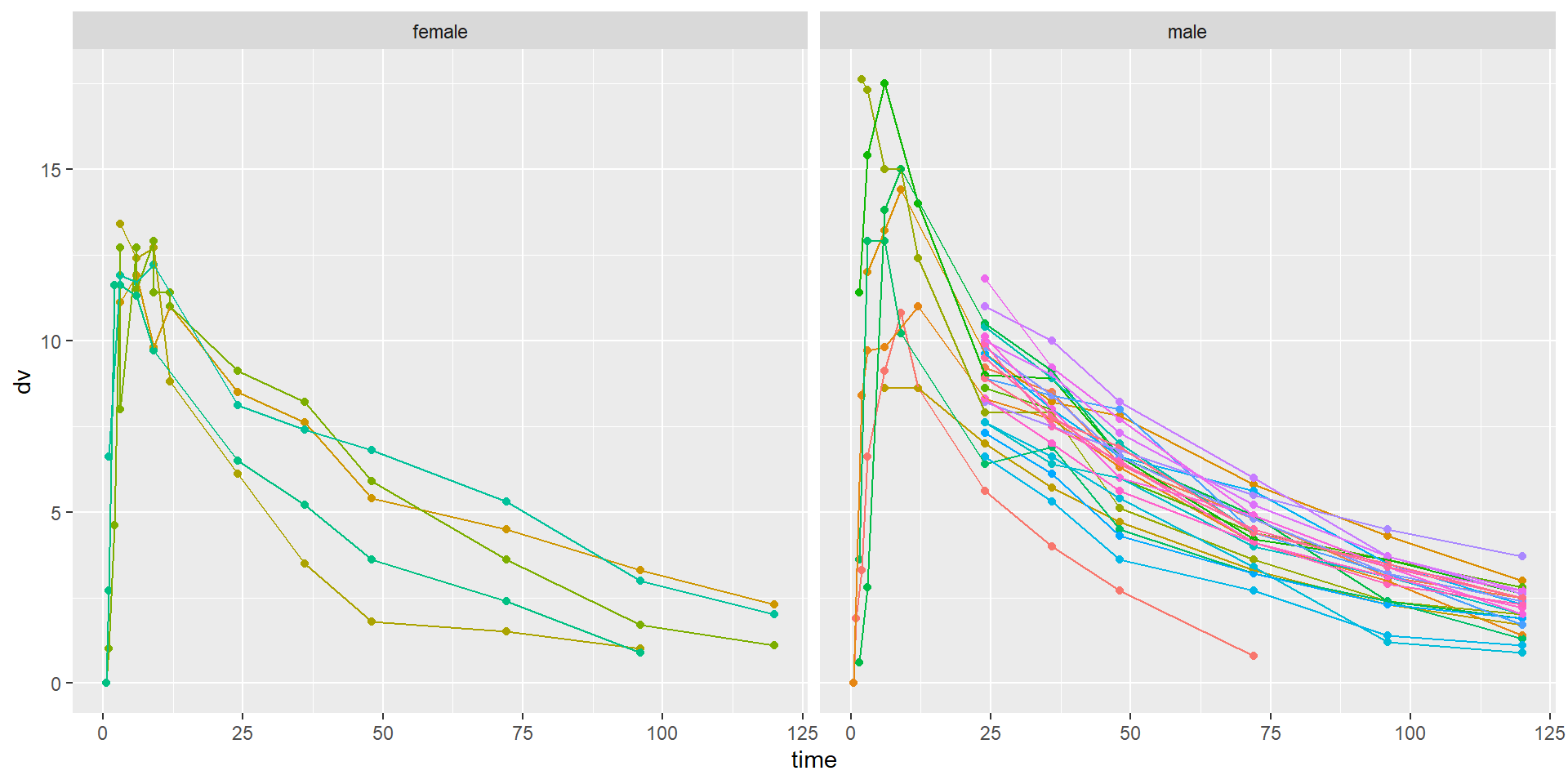
Min. :0 Min. : 40.00 Min. :21.00 female: 57 low wt :133
1st Qu.:0 1st Qu.: 59.00 1st Qu.:23.00 male :194 high wt:118
Median :0 Median : 70.00 Median :31.00
Mean :0 Mean : 68.71 Mean :32.47
3rd Qu.:0 3rd Qu.: 78.00 3rd Qu.:40.00
Max. :0 Max. :102.00 Max. :63.00
age_cat
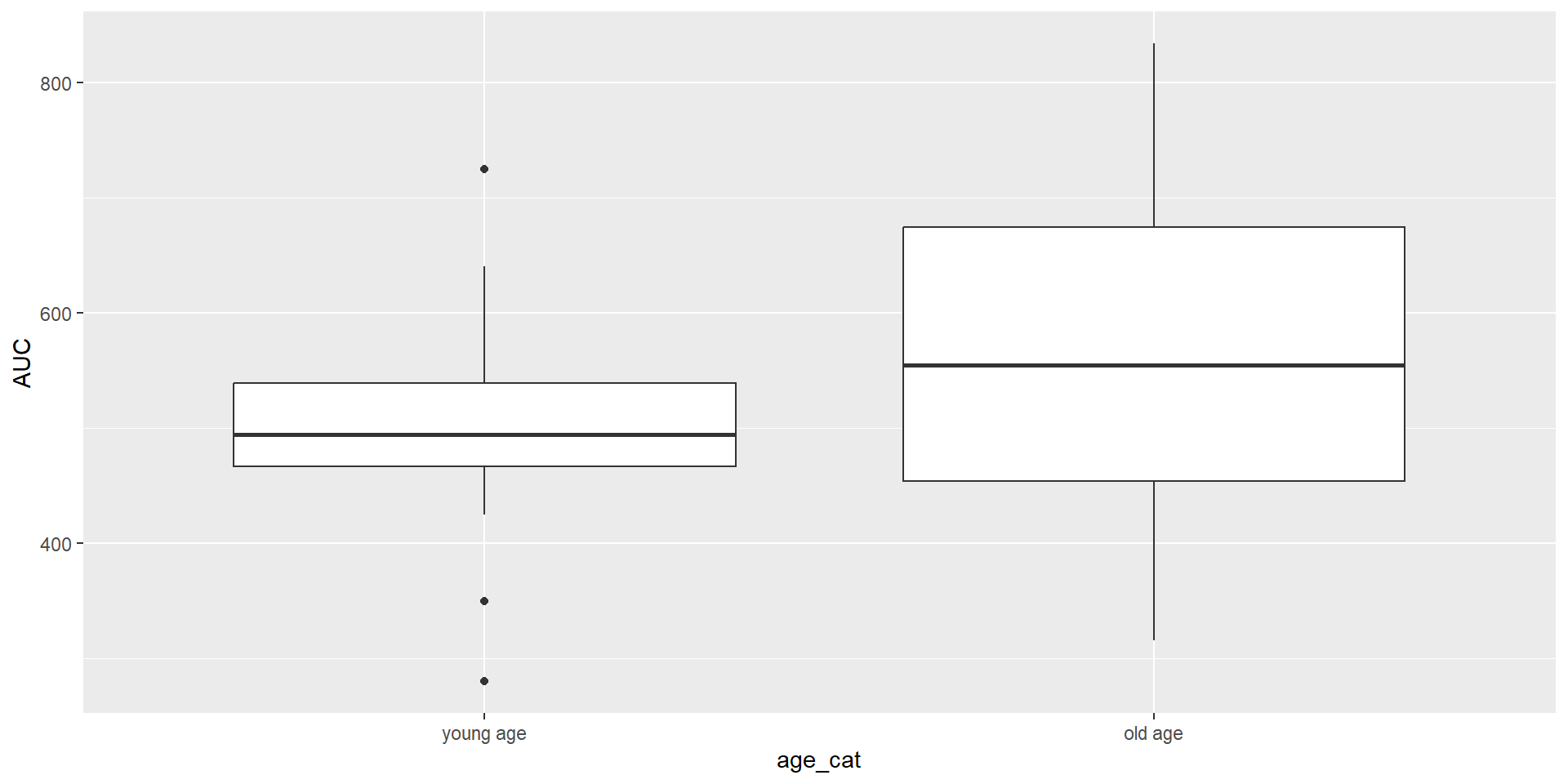
young age:123
old age :128